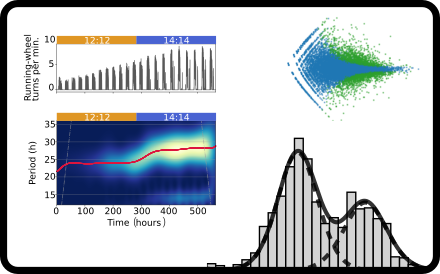
Data science
We use methods from bioinformatics, image processing, time series analysis and statistics to test for rhythmic patterns in data and to infer oscillatory parameters such as periods, amplitudes and phases.

We use methods from bioinformatics, image processing, time series analysis and statistics to test for rhythmic patterns in data and to infer oscillatory parameters such as periods, amplitudes and phases.

Delayed negative feedback loops convergentlly evolved as a mechanism underlying circadian rhythm generation across different kingdoms of life. We study design principles of these motifs using data driven modeling and dynamical systems theory.

Intercellular communication and emergent network properties are quintessential for circadian system level functioning. We use spatial statistics and simulations of oscillator networks to characterize spatial patterns at the tissue and organ level.
Stable entrainment of the intrinsic circadian clock to 24h environmental rhythms is of fundamental importance for the proper alignment of physiological processes around the solar day and is thus under evolutionary selection. We use concepts from oscillator theory and simulations of molecular clock models to investigate design principles underlying experimentally observed entrainment characteristics under laboratory and natural conditions.